This is the story of my journey through my research, to Peppytide – and Physical Scientific Computing. It’s a history of how it all started.
This is the story of my journey through my research, to Peppytide – and Physical Scientific Computing. It’s a history of how it all started.
This photo was taken at the time I first made the model work in the summer of 2011 with all the sidechains. This was just the backbone chain at first to check how the dynamics of the main chain would work out, in terms of folding into secondary structures. After that, it was time to plug-in the side chains, which ya’ll know came out spectacular in red and blue.
Before this model, I had another version with just the backbone, with the side-chains being represented by place-holding plastic and/or polystyrene balls, whose radius was in accordance with the calculated sum-radius of the cluster of side-chain atoms taken together.
Here are some photos of that version, along with it being folded into an alpha-helix for the first time. But the steric interaction of the model made it hard to fold into the helix. And it was pushing as well, so some hydrogen-bonds were opening up, so the helix was not so stable. It was apparent that the side chain atoms, when I do make them, would be a snug fit. This was exciting.




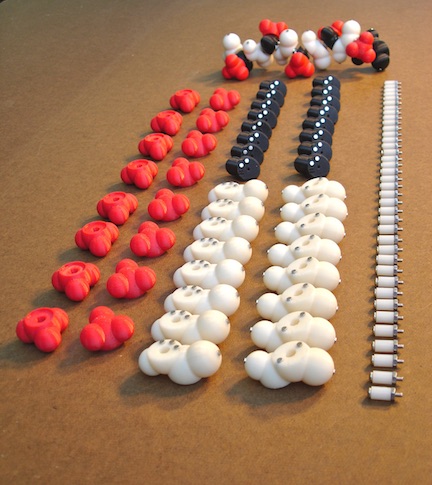
But the really really absolutely first 3D prints were these parts below, that I printed at UCSF. This does look so different from what it ultimately became.

So, why make such a model? Here goes the story.
I see the world in terms of motion – atoms moving around in the many ways of nature. I can remember this particular way of observing the world around, as far back as I can tunnel into my childhood.
I love counting and measuring things as well – countably finite, countably infinite, uncountably infinite, just infinite, measure for n-dimensional spaces, counting of things in motion, vectors, curls, and many more ways of permutations and combinations of these countings. In short, along with the science of nature, I love mathematics too. I studied Mathematics major for my undergrads in Kolkata, India, with minors in Physics and Chemistry. My love for counting all things logical, practical and natural led to my affinity for Computer Science. So simultaneously with the regular Mathematics degree (BS), I did a correspondence Bachelor’s degree in Computer Science (BCA, computer science and applications) from a distance-learning university in India. Then a full-time masters in Computer Science (MCA) in Kolkata, before immigrating to United States, where at LSU I did another master’s in CS with a focus on Grid Computing, HPC, computational biology and molecular dynamics, and ultimately a PhD in Computer Science from Virginia Tech with a focus in protein dynamics and modeling. So a lot of science, math and degrees there. I am what is called a hybrid – an interdisciplinary researcher – and I love all of it.
Before coming to USA, I worked briefly as a researcher at the Indian Statistical Institute, Kolkata, (ISI), a top research institute in statistics and computer science, where I worked in Algorithms and Data Structure in my first year, and GPS positioning algorithms and cellular networking in my second year. In addition, I was spending my home-time doing small projects in and learning more about AI and Neuroinformatics. Yes I have a thing for the brain! I am now making a brain model. In addition to learning all these new things, I was of course studying for GRE, TOEFL, and trying to learn how to apply to US universities. After ISI, I worked a few short months as a software developer in Siemens, Kolkata, working in cellular networking for their off-shore client. Hence C++ was lunch and dinner at that time.
When I moved to LSU in USA in the fall of 2005, all the excitement was at the Center for Computation and Technology (CCT) in High Performance Computing (HPC) and computational science. Mixing that with the color of my background, I ended up studying molecular dynamics and parallel algorithms for delving into amino acids and proteins. The run-time execution of these codes being spread across multiple supercomputers as testbeds all over the nation. This work was for my masters (MS). You can read one of my papers for this work here published at the Philosophical Transactions of the Royal Society A.
I moved on to Virginia Tech (VT) for a PhD in Computer Science in fall of 2008. The department is known for being open to interdisciplinary approaches – and I was indeed tantalized by these interdisciplinary possibilities. The first year, I spent working in an app for Cell Cycle called Jigcell. Next year, with the promise of more independence for my dream project of developing macromolecules in motion, I moved to another lab that promised independence.
I spent the summer of 2009 doing an internship at the Berkeley Lab, getting my hands dirty with the supercomputers that were then housed at the Oakland Scientific Facility.
Back to VT in Fall, over the next two years (2009 – 2011, May), I conceptualized several versions of digital-physical systems for amino acid molecules, nucleotides and proteins – the work that would ultimately culminate, a few years later, into Peppytide and Physical Biomoldeing and then to Physical Scientific Computing over multiple years. In the summer of 2011, I (again) moved on to Berkeley Lab’s Material Sciences Division (Molecular Foundry, Biological Nanostructures) in search of more independence and more ‘interdisciplinarity’.
One of the systems that I had conceptualized at VT was focusing on the Central Dogma of biology. I named it ‘BioTable’ – partly digital and partly a mock physical system with wood, paper and what-not to prototype the concept. And it had to be very very cheap, and hence the choice of materials.
Yes that table in the photo below is a computer monitor. The software that I had developed was used to work with these paper/wood/bead models. I was focussing on both tRNA folding and protein folding.
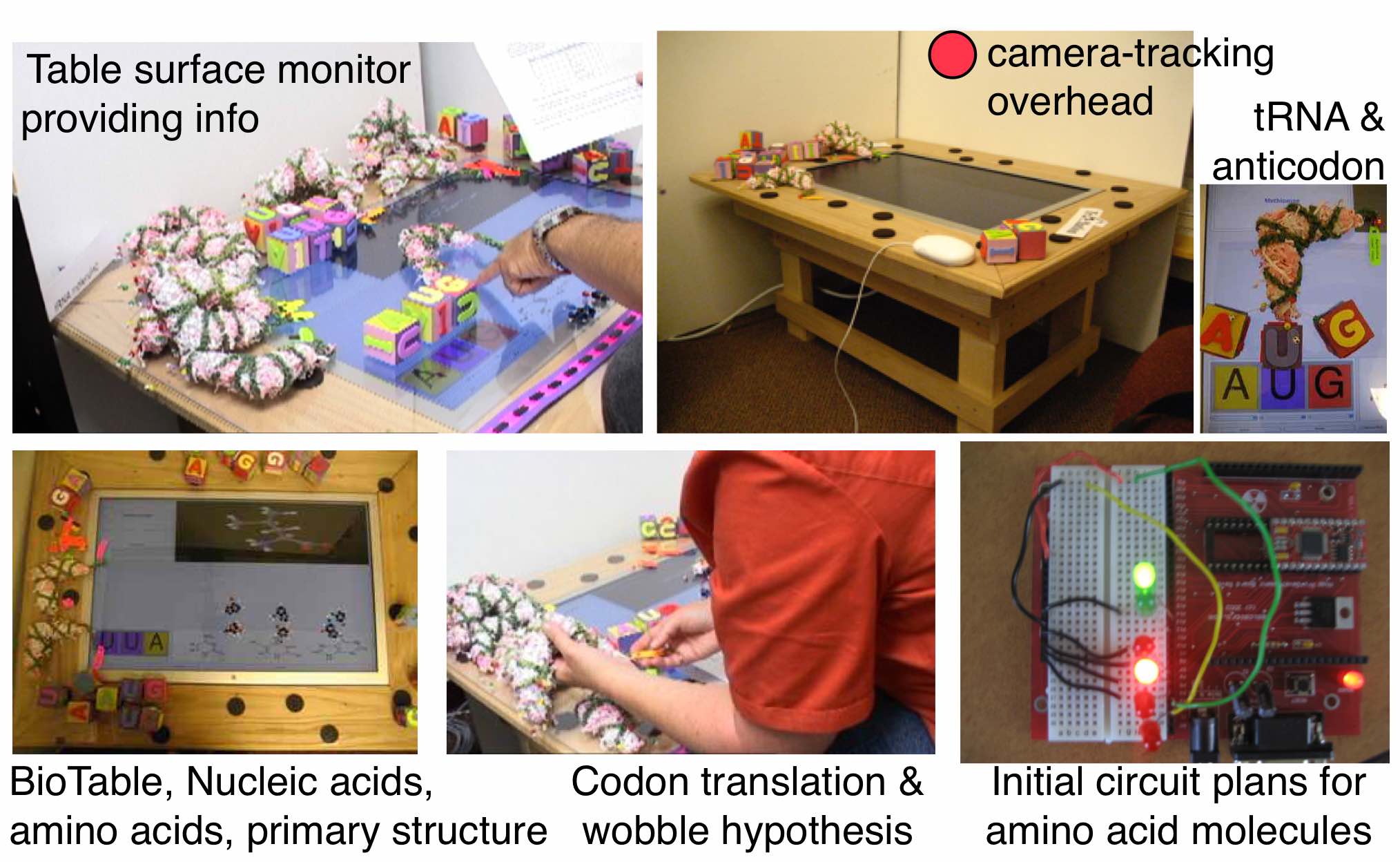
Fig: BioTable
Some nucleotides and hydrogen-bonds, for DNA (yes I made them with beads, copper and crystals):
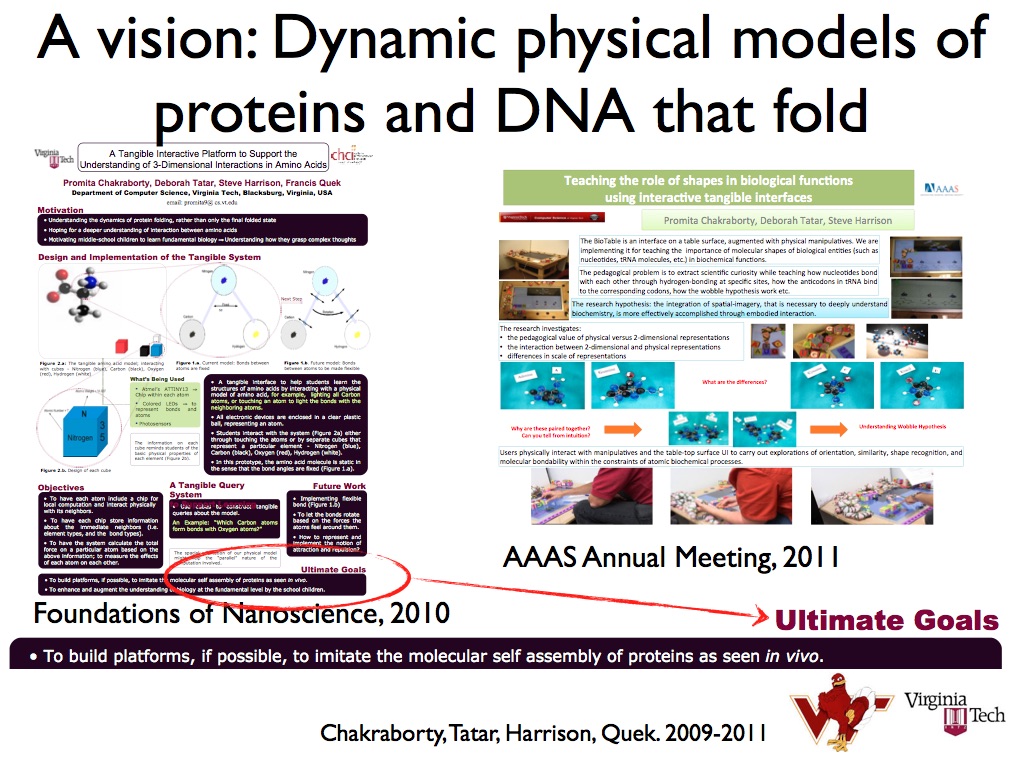
Another part of the project I conceptualized at the same time, was to study the forces experienced by different amino acid molecules and shifts in their bond angles as they were brought closer to each other physically. Here is a partial glimpse of the poster I presented for that:
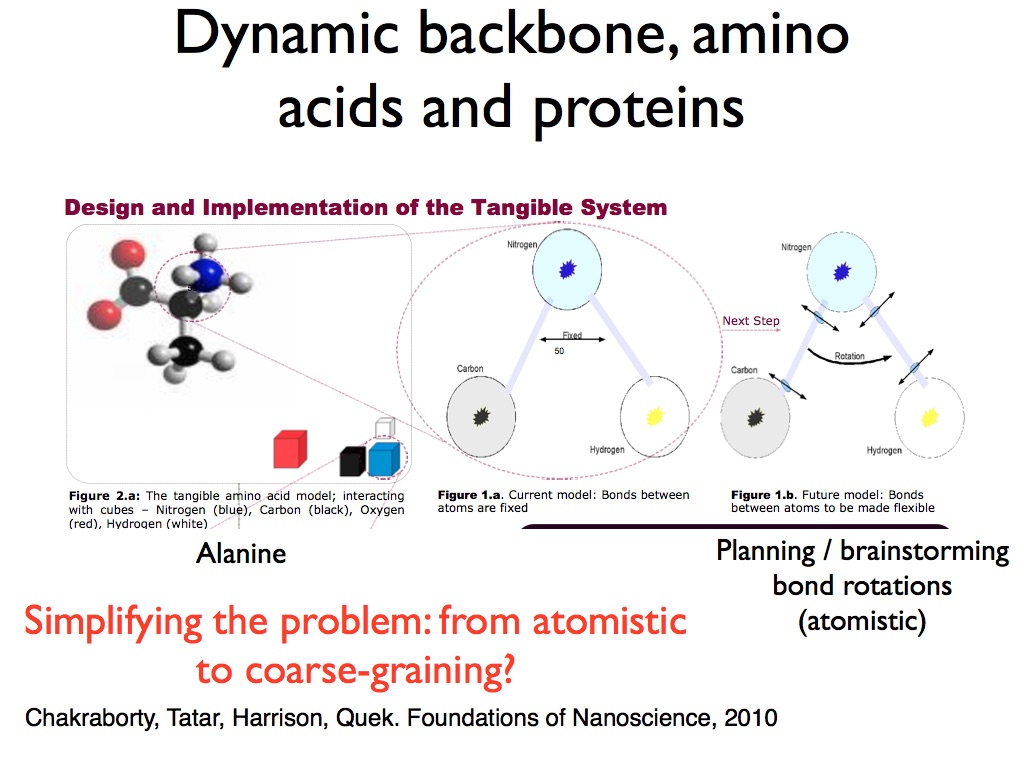
I presented a few posters of these concepts in various conferences before joining Berkeley Lab in 2011. I presented at these conferences : Foundations of Nanoscience 2010, Grace Hopper Celebration 2010, AAAS Annual Meeting 2011. Some photos of the various posters:
I felt like I needed a 3D Printer for the ultimate prototyping, and some CAD algorithms. I spent the Spring of 2011 at VT learning to draw CAD models using Google SketchUp, Autodesk Inventor and Solidworks. I took some available training sessions as well for Inventor. I also explored and looked at some 3D printers that were available and functional at the Mechanical Sciences department at VT.
All these mini-projects were branches of the same problem that I was attacking from several angles: to design for a system of physical dynamic models of macromolecules like proteins, RNA and DNA that would exhibit their respective, idiosyncratic molecular dynamics, and would be measurable by hand+computer in some way. Thus, by the time I joined LBL, through my unique journey, I had accumulated all the background that I needed to make my vision a reality – quite a laundry list of things: Algorithms, Computational Biology, Scientific Computing, 3D Mathematical Modeling, Visualizing, Big data, Structural biology, HPC, Grid (the ancestor of Cloud Computing), CAD algorithms, HCI, Deep learning, Data Science and Math.
I joined Berkeley Lab in May 2011, with my vision traveling with me. It must have been an immense stroke of luck – or so I thought at the moment – that I was given all the independence that I needed to develop my models. I came as a student research intern to Berkeley Lab, but after the successful demonstration, I was hired as a full time term employee as a Research Associate in January 2012. A 3D printer was also bought next January (2012) at the Molecular Foundry, after I had demonstrated that the initial version worked, using a 3D Printer at UCSF’s Prof. Joe DeRisi’s lab user facilities. Joe loved my models and would show them off around the halls of UCSF to everyone. I thank him so much for his help and encouragement.
I have worked in a couple of startup workspaces and used their labs and machines. Now I have two 3d printers set up in my garage workshop at home, complete with drills and other tools. I experiment for new models (of brain) here, and also for next versions of Peppytides.
From May through July 2011, I went through rounds of designing, 3D printing and machining of the protein model. With each passing round, the accuracy improved. By end of summer, I had the first prototype in hand. I coined the term “Peppytide” around that time; here’s the etymology and reasons: (1) the word “peppy” means lively, (2) the word “peptide” is small polypeptide chains, (3) started with P (my name’s initial).

Fall 2011 passed by with building improved versions – lighter, more accurate, more flexible. I arrived at the somewhat final version in summer of 2012. I presented a poster about it at the Foundry Annual User Meeting in the Fall of 2012. I was so surprised to get the Best Poster Award, among all amazing works of Foundry users’ research and materials sciences research. I defended and passed my PhD proposal with the model in Virginia Tech Computer Science department in May 2012.
I published the design and details of the scientific accuracy of Peppytide in a paper in PNAS (Proceedings of National Academy of Sciences) in Jul 2013, Make Magazine (Jan 2014) and PhD dissertation (Spring 2014). A list of my publications and talks is in my homepage publications list.
By the time I left Berkeley Lab in July 2014, with the Director’s Award in tow, Physical Biomodeling as a possible new field of study was firmly established in my mind, and that had reflected in my dissertation as well. I summarized the concepts in an arxiv paper (Feb 2015).
Physical Biomodeling eventually extended into a broader vision of Physical Scientific Computing, not just for biology.
By then it had taken a decade of study and experimentation to come to that point. To think that a decade’s worth of study and philosophizing can be zipped into a paper so short!
I presented the gist of that paper in AAAS Annual Meeting 2015. Here’s a slide:
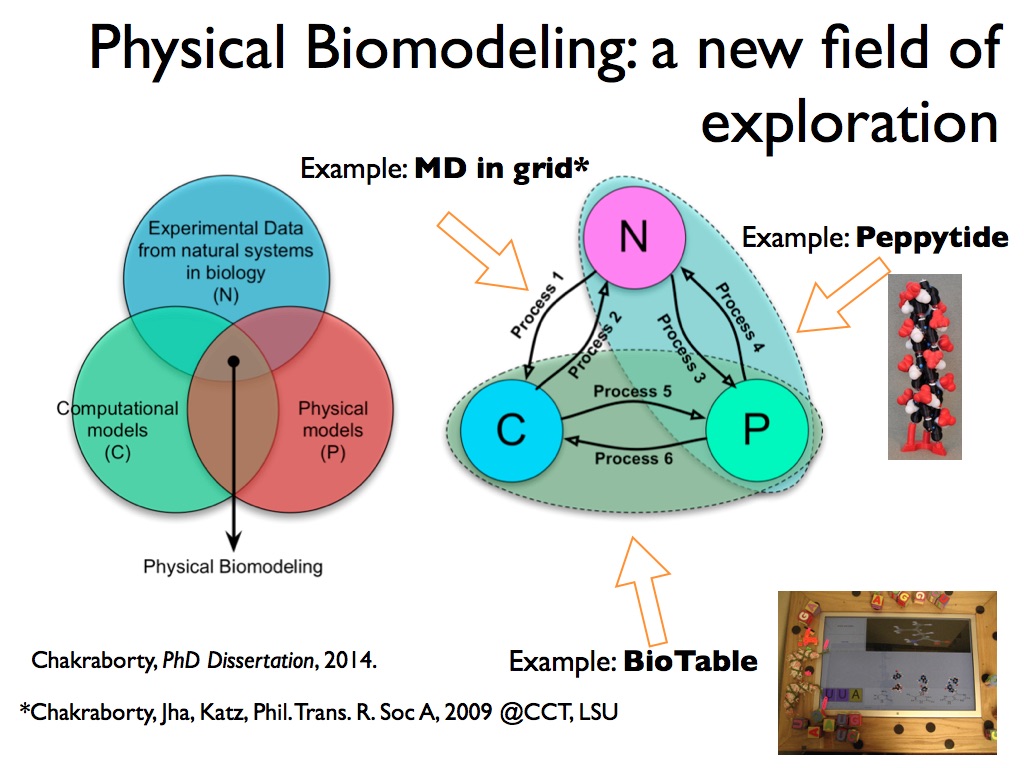
Fig: A slide from my talk at AAAS Annual Meeting 2015 (entire presentation here in SlideShare)
Peppytide is just the tip of the iceberg of that culmination. There are many more projects to come. One project that I am currently conceptualizing – that would not see the daylight for years – is a physical, flexible, measurable model for the human brain. Yes it has to come to that! And yes, I work slow and steady! Another work-in-progress project is a model of the eye.
Physical Scientific Computing as a field is here to stay.
There are of course so many people who have joyfully experienced my work. A very, very tiny glimpse:

I leave you with this photo of the parts that I took wayyyy back in 2011. It is a metaphor for my journey as a scholar: the parts don’t always show how the end-product would look like, and the end doesn’t show the story of its constituents. Especially when something new is being cooked in the covered cauldron.